Plot 3D Effects
plot3d.RdFunction to plot 3D graphics or image and/or contour plots for bivariate effects/functions.
plot3d(x, residuals = FALSE, col.surface = NULL,
ncol = 99L, swap = FALSE, col.residuals = NULL, col.contour = NULL,
c.select = NULL, grid = 30L, image = FALSE, contour = FALSE,
legend = TRUE, cex.legend = 1, breaks = NULL, range = NULL,
digits = 2L, d.persp = 1L, r.persp = sqrt(3), outscale = 0,
data = NULL, sep = "", shift = NULL, trans = NULL,
type = "mba", linear = FALSE, extrap = FALSE,
k = 40, ...)Arguments
- x
A matrix or data frame, containing the covariates for which the effect should be plotted in the first and second column and at least a third column containing the effect. Another possibility is to specify the plot via a
formula, e.g. for simple plotting of bivariate surfacesz ~ x + y, see the examples.xmay also be a character file path to the data to be used for plotting.- residuals
If set to
TRUE, residuals may also be plotted if available. Residuals must be supplied as anattribute named"residuals", which is a matrix or data frame where the first two columns are covariates and the third column the residuals.- col.surface
The color of the surface, may also be a function, e.g.
col.surface = heat.colors.- ncol
the number of different colors that should be generated, if
col.surfaceis a function.- swap
If set to
TRUEcolors will be represented in reverse order.- col.residuals
The color of the partial residuals, or if
contour = TRUEthe color of the contour lines.- col.contour
The color of the contour lines.
- c.select
Integer vector of maximum length of columns of
x, selects the columns of the resulting data matrix that should be used for plotting. E.g. ifxhas 5 columns, thenc.select = c(1, 2, 5)will select column 1, 2 and 5 for plotting. Ifc.select = 95orc.select = 80, functionplot3dwill search for the corresponding columns to plot a 95\(\%\) or 80\(\%\) confidence surfaces respectively. Note that if e.g.c.select = c(1, 2),plot3dwill use columns 1 + 2 and 2 + 2 for plotting.- grid
The grid size of the surface(s).
- image
If set to
TRUE, animage.plotis drawn.- contour
If set to
TRUE, acontourplot is drawn.- legend
If
image = TRUEan additional legend may be added to the plot.- cex.legend
The expansion factor for the legend text, see
text.- breaks
A set of breakpoints for the colors: must give one more breakpoint than
ncol.- range
Specifies a certain range values should be plotted for.
- digits
Specifies the legend decimal places.
- d.persp
See argument
din functionpersp.- r.persp
See argument
rin functionpersp.- outscale
Scales the outer ranges of
xandzlimits used for interpolation.- data
If
xis aformula, adata.frameorlist. By default the variables are taken fromenvironment(x): typically the environment from whichplot3dis called. Note thatdatamay also be a character file path to the data.- sep
The field separator character when
xordatais a character, see functionread.table.- shift
Numeric constant to be added to the smooth before plotting.
- trans
Function to be applied to the smooth before plotting, e.g., to transform the plot to the response scale.
- type
Character, which type of interpolation method should be used. The default is
type = "akima", see functioninterp. The two other options aretype = "mba", which calls functionmba.surfof package MBA, ortype = "mgcv", which uses a spatial smoother withing package mgcv for interpolation. The last option is definitely the slowest, since a full regression model needs to be estimated.- linear
Logical, should linear interpolation be used withing function
interp?- extrap
Logical, should interpolations be computed outside the observation area (i.e., extrapolated)?
- k
Integer, the number of basis functions to be used to compute the interpolated surface when
type = "mgcv".- ...
Parameters passed to
colorlegendif an image plot with legend is drawn, also other graphical parameters, please see the details.
Details
For 3D plots the following graphical parameters may be specified additionally:
cex: Specify the size of partial residuals,col: It is possible to specify the color for the surfaces ifse > 0, then e.g.col = c("green", "black", "red"),pch: The plotting character of the partial residuals,...: Other graphical parameters passed functionspersp,image.plotandcontour.
Note
Function plot3d can use the akima package to construct smooth interpolated
surfaces, therefore, package akima needs to be installed. The akima package has an ACM
license that restricts applications to non-commercial usage, see
https://www.acm.org/publications/policies/software-copyright-notice
Function plot3d prints a note referring to the ACM license. This note can be suppressed by
setting
options("use.akima" = TRUE)
See also
Examples
## Generate some data.
set.seed(111)
n <- 500
## Regressors.
d <- data.frame(z = runif(n, -3, 3), w = runif(n, 0, 6))
## Response.
d$y <- with(d, 1.5 + cos(z) * sin(w) + rnorm(n, sd = 0.6))
if (FALSE) ## Estimate model.
b <- bamlss(y ~ s(z,w), data = d)
summary(b)
#> Error in eval(expr, envir, enclos): object 'b' not found
## Plot estimated effect.
plot(b, model = "mu", term = "s(z,w)")
#> Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'plot': object 'b' not found
## Extract fitted values.
f <- fitted(b, model = "mu", term = "s(z,w)", intercept = FALSE)
#> Error in eval(expr, envir, enclos): object 'b' not found
f <- cbind(d[, c("z", "w")], f)
#> Error in eval(expr, envir, enclos): object 'f' not found
## Now use plot3d().
plot3d(f)
#> Error in eval(expr, envir, enclos): object 'f' not found
plot3d(f, swap = TRUE)
#> Error in eval(expr, envir, enclos): object 'f' not found
plot3d(f, grid = 100, border = NA)
#> Error in eval(expr, envir, enclos): object 'f' not found
## Only works if columns are named with
## '2.5
plot3d(f, c.select = 95, border = c("red", NA, "green"),
col.surface = c(1, NA, 1), resid = TRUE, cex.resid = 0.2)
#> Error in eval(expr, envir, enclos): object 'f' not found
## Now some image and contour.
# plot3d(f, image = TRUE, legend = FALSE)
# plot3d(f, image = TRUE, legend = TRUE)
# plot3d(f, image = TRUE, contour = TRUE)
# plot3d(f, image = TRUE, contour = TRUE, swap = TRUE)
# plot3d(f, image = TRUE, contour = TRUE, col.contour = "white")
# plot3d(f, contour = TRUE)
# plot3d(f, image = TRUE, contour = TRUE, c.select = 3)
# plot3d(f, image = TRUE, contour = TRUE, c.select = "Mean")
# plot3d(f, image = TRUE, contour = TRUE, c.select = "97.5
## Variations.
d$f1 <- with(d, sin(z) * cos(w))
with(d, plot3d(cbind(z, w, f1)))
## Same with formula.
plot3d(sin(z) * cos(w) ~ z + w, zlab = "f(z,w)", data = d)
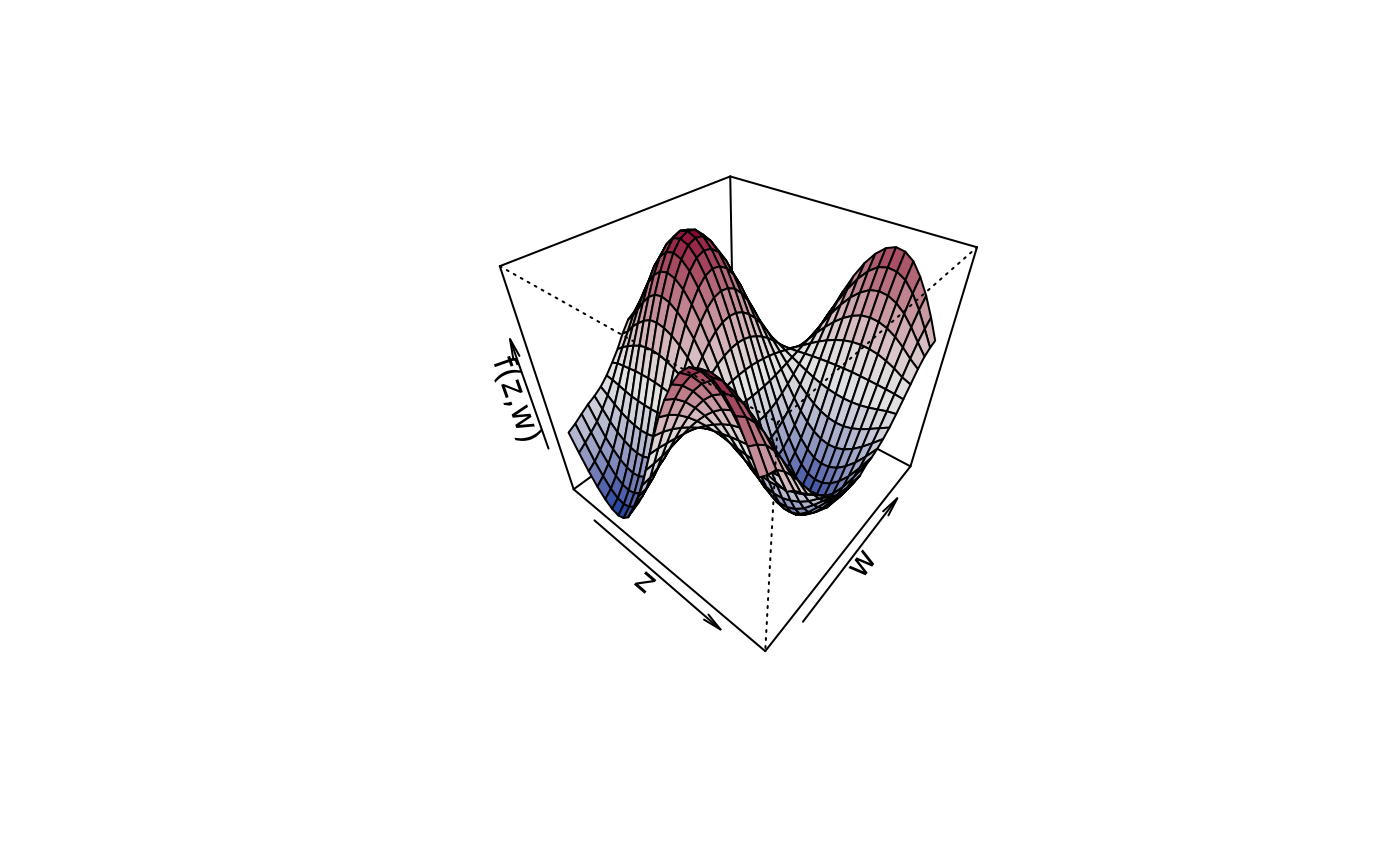 plot3d(sin(z) * cos(w) ~ z + w, zlab = "f(z,w)", data = d,
ticktype = "detailed")
plot3d(sin(z) * cos(w) ~ z + w, zlab = "f(z,w)", data = d,
ticktype = "detailed")
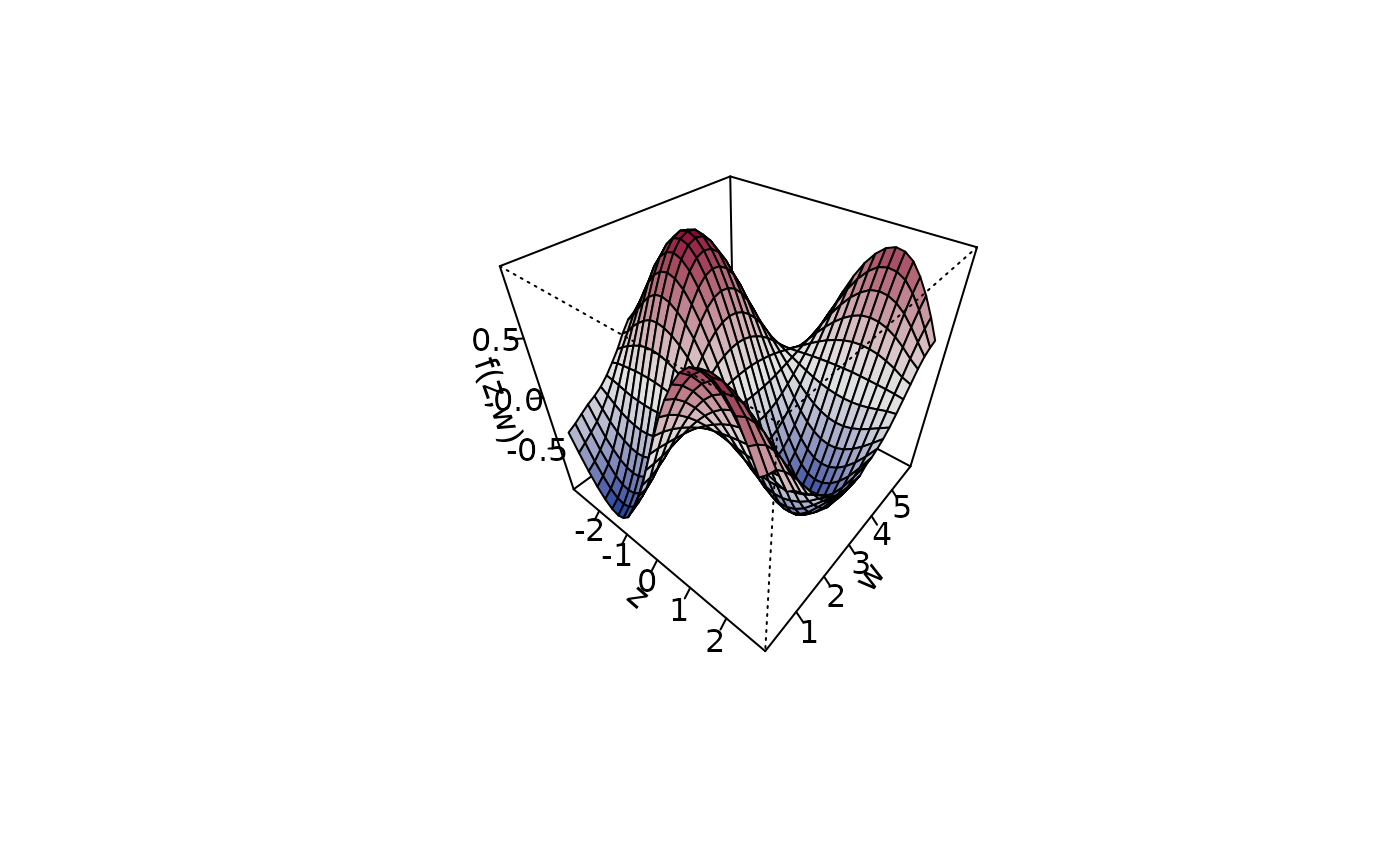 ## Play with palettes.
plot3d(sin(z) * cos(w) ~ z + w, col.surface = heat.colors, data = d)
## Play with palettes.
plot3d(sin(z) * cos(w) ~ z + w, col.surface = heat.colors, data = d)
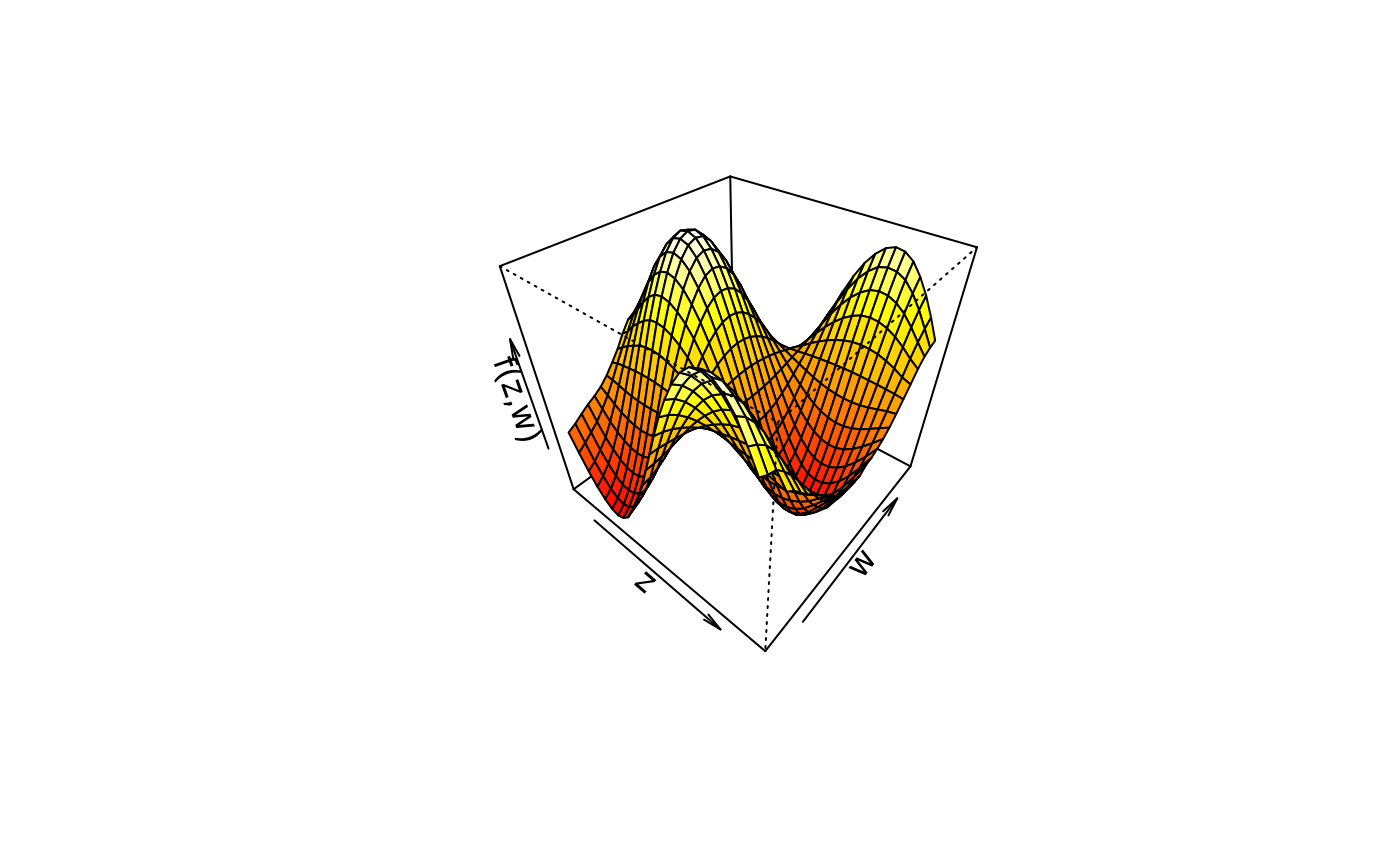 plot3d(sin(z) * cos(w) ~ z + w, col.surface = topo.colors, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = topo.colors, data = d)
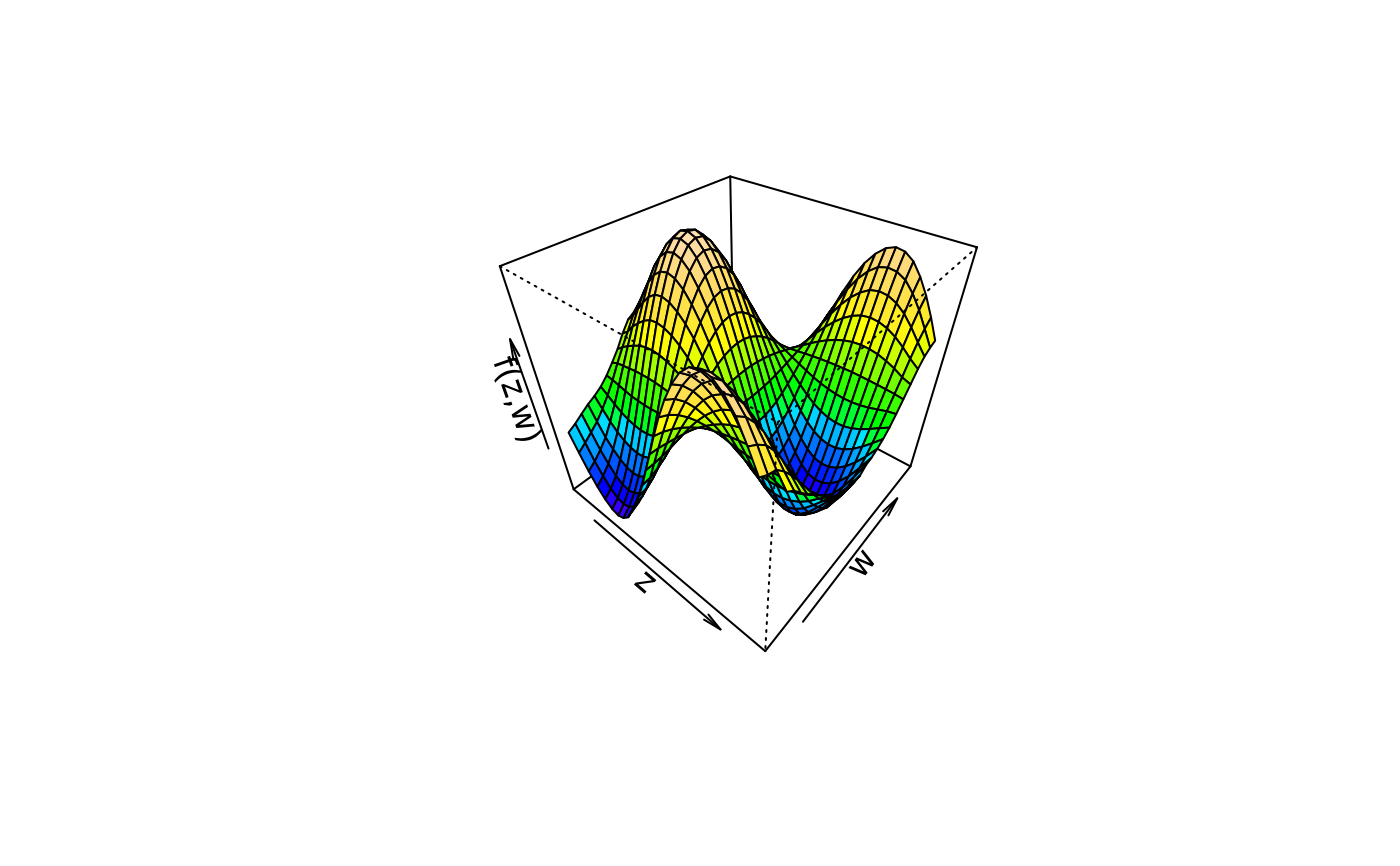 plot3d(sin(z) * cos(w) ~ z + w, col.surface = cm.colors, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = cm.colors, data = d)
 plot3d(sin(z) * cos(w) ~ z + w, col.surface = rainbow, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = rainbow, data = d)
 plot3d(sin(z) * cos(w) ~ z + w, col.surface = terrain.colors, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = terrain.colors, data = d)
 plot3d(sin(z) * cos(w) ~ z + w, col.surface = rainbow_hcl, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = rainbow_hcl, data = d)
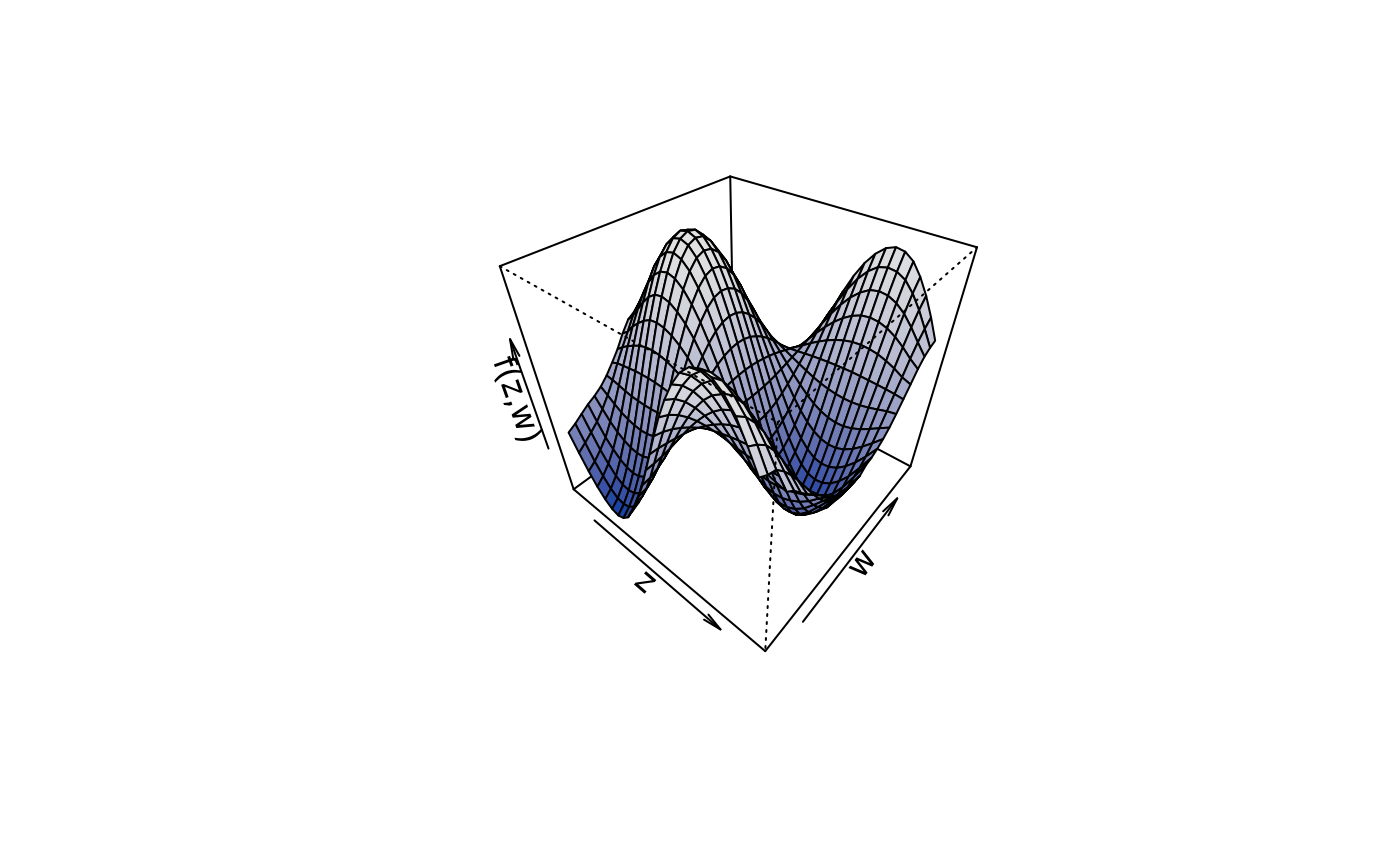
 plot3d(sin(z) * cos(w) ~ z + w, col.surface = diverge_hcl, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = diverge_hcl, data = d)
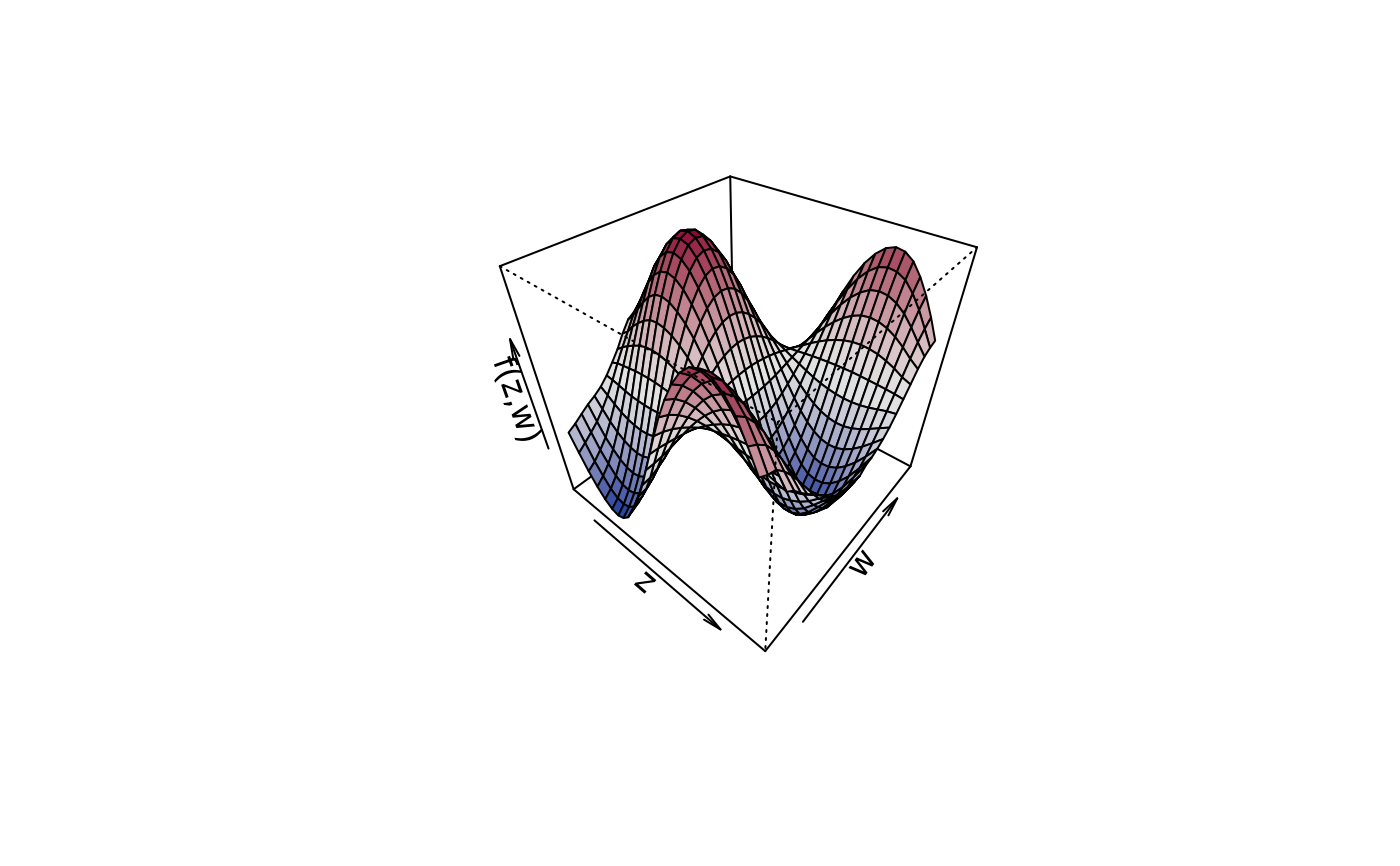 plot3d(sin(z) * cos(w) ~ z + w, col.surface = sequential_hcl, data = d)
plot3d(sin(z) * cos(w) ~ z + w, col.surface = sequential_hcl, data = d)
 plot3d(sin(z) * cos(w) ~ z + w,
col.surface = rainbow_hcl(n = 99, c = 300, l = 80, start = 0, end = 100),
data = d)
plot3d(sin(z) * cos(w) ~ z + w,
col.surface = rainbow_hcl(n = 99, c = 300, l = 80, start = 0, end = 100),
data = d)
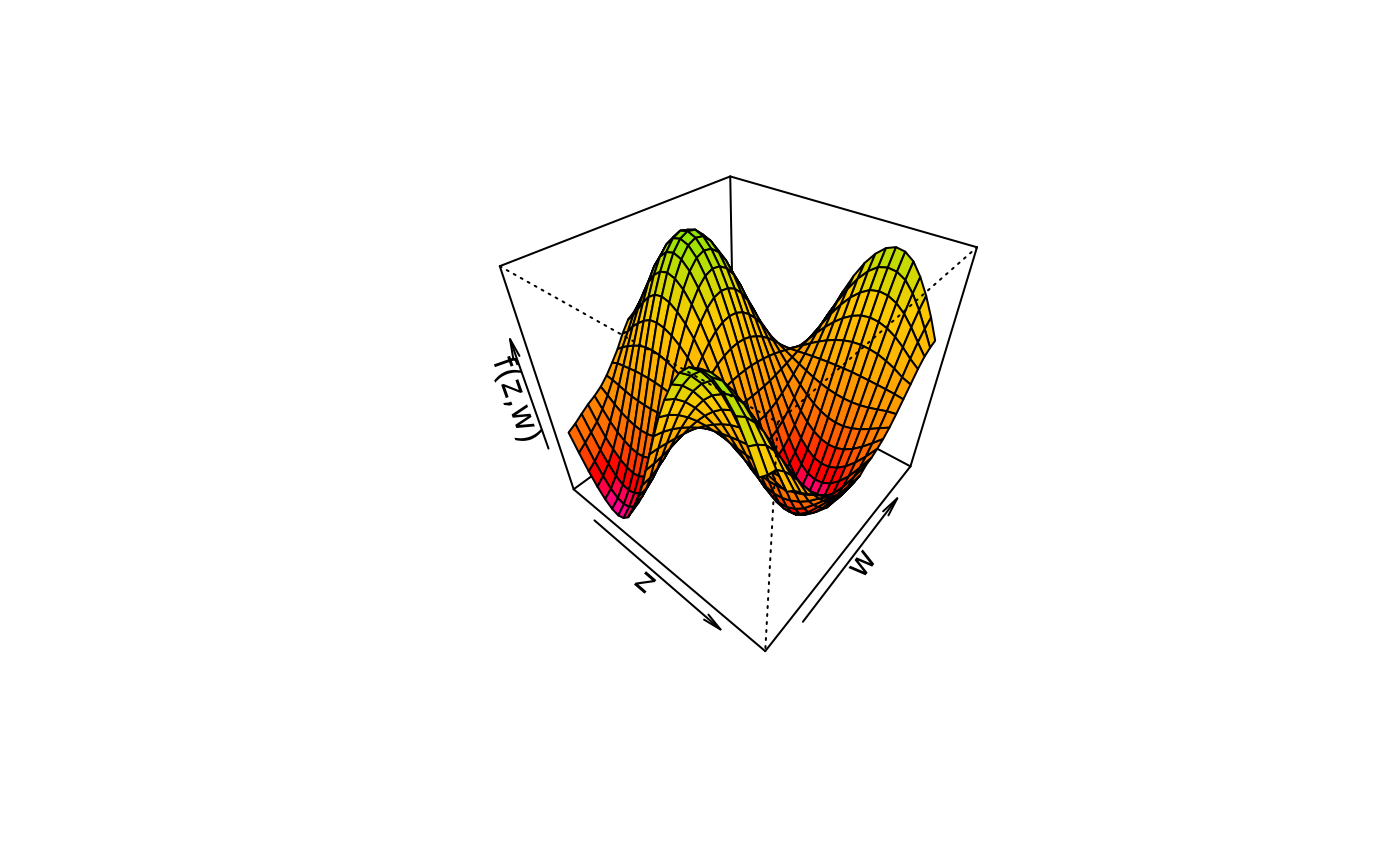 # plot3d(sin(z) * cos(w) ~ z + w,
# col.surface = rainbow_hcl(n = 99, c = 300, l = 80, start = 0, end = 100),
# image = TRUE, grid = 200, data = d)
# plot3d(sin(z) * cos(w) ~ z + w,
# col.surface = rainbow_hcl(n = 99, c = 300, l = 80, start = 0, end = 100),
# image = TRUE, grid = 200, data = d)